Computer-aided molecular design (CAMD) emerged from advances in computational chemistry and computer technology, and promises to revolutionize the design of functional molecules. In the pharmaceutical industry, CAMD aims to identify highly potent and specific drugs using computational methods and structural information on the target biomolecules. Our research activities in this area focus on the development of new computational tools for CAMD and their application to the design of new drugs and other nano-scale molecular devices
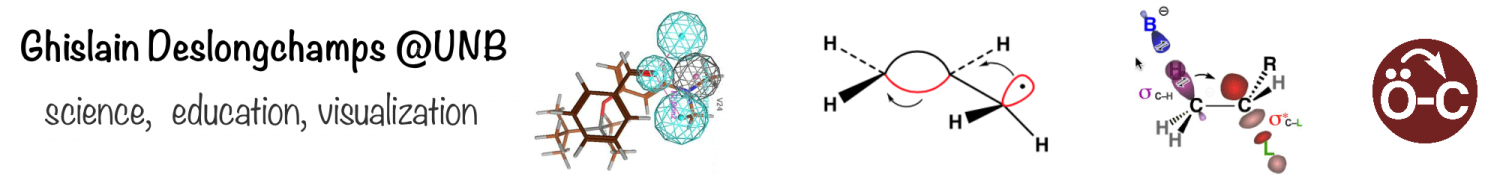
Reverse-docking: We have developed “reverse-docking” as a new paradigm for the computer-assisted design of artificial receptors, chemosensors, catalysts and other nano-molecular devices. The basic approach hinges on docking large, flexible molecular receptors/catalysts around rigid ligands/transition state models. Using our reverse-docking program EM-Dock, we have demonstrated its application for the study of asymmetric organocatalysts, metal-free catalysts capable of directing asymmetric reactions. The resulting 3D models of organocatalyzed reactions provide insight into the mode of asymmetric induction, and predict the product enantiomers in 100% of cases studied to date. Efforts are currently underway to broaden the scope of reverse-docking applications, including the virtual screening of asymmetric catalysts. Our EM-Dock was written in SVL for integration into the Molecular Operating Environment (MOE) drug discovery platform. It is freely available through the SVL Exchange.
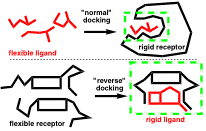
GI-MOE: GI-MOE (Gaussian Interface for MOE) is a full-featured interface for carrying out large-scale quantum mechanical calculations within MOE using Gaussian on the Compute Canada advanced research computing (ARC) facility. GI-MOE automates the calculations on large databases of molecules, including geometry optimizations, transition-state calculations, potential scans, as well as ONIOM calculations. It generates the Gaussian .com files and the SLURM shell files required for running Gaussian on the Compute Canada facility. GI-MOE was written in SVL for integration into MOE and is freely available through the SVL Exchange.